Interdisciplinary research with Dr. Rebeca Davis and Dr. Pingzhao Hu and their lab members
Prediction of antibiotic activity based on chemogenomic profiles
We are building bacterial knockdown libraries as genetic tools to predict antibiotic activity in new compounds and to determine their mechanism of action. Our collections contain knockdown mutants in essential genes, which usually code for targets of antibiotics. The overarching hypothesis of our research is that chemogenetic profiles of essential gene knockdowns will better predict antibiotic activity and mode of action (MOA) of novel compounds, accelerating antibiotic discovery. Our knockdown CRISPRi libraries are valuable tools for machine learning-based prediction of antibiotic activity and mode of action of any active antimicrobial compound. The knockdown mutants have different susceptibility to molecules with antibiotic activity, which can signal drug-target interactions.
Drug-gene interaction matrices can help us develop a machine learning platform (MLP) that can be used for in silico screening of ultra-large, chemically diverse virtual libraries to accurately predict the bioactivity and MOA of the compounds. This approach will unlock the possibilities of virtually testing billions of ‘drug-like’ compounds and exploration of broader targets (essential genome) and chemical scaffolds that are not available otherwise and thus can increase the probability of finding new drug classes.
Link to article: Deep Learning-Driven prediction of drug mechanism of action from large-scale chemical-genetic interaction profiles by Chengyou Liu et al., J Chemoinformatics 2022.

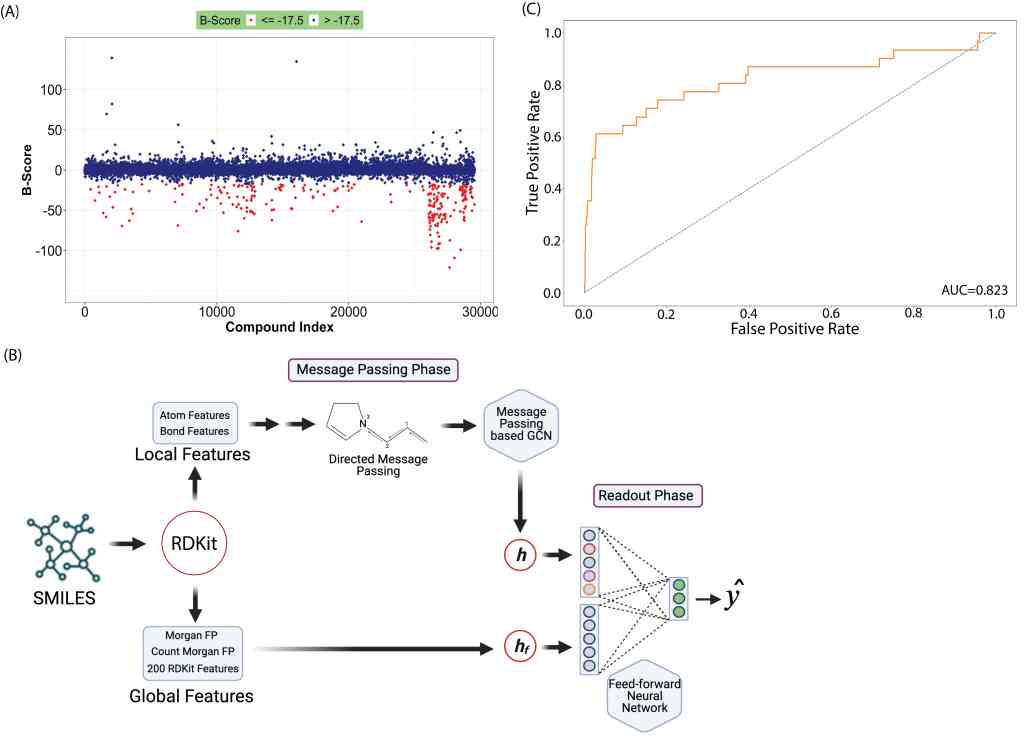
Using AI to predict antibiotic activity in small molecules virtual libraries
Screens for antibacterial activity in small molecule libraries have very little success. We used a previosuly developed deep learning approach, and a former screen of 30,000 compounds for growth inhibitory activity against the antibiotic-resistant bacterium Burkholderia cenocepacia. We trained the ML modelto achieve a receiver operating characteristic (ROC) score of 0.823 on the test set. We predicted antibacterial activity in virtual libraries and tested the predicted compounds in the laboratory. More than 50% of the compounds with growth inhibitory activiy inhibited ESKAPE pathogens. Overall, the developed ML approach can be used for compound prioritization before screening, increasing the typical hit rate of drug discovery.
Link to article: A machine learning model trained on a high-throughput antibacterial screen increases the hit rate of drug discovery by Zisan Rahman et al., PLOS Computational Biology 2022
Funding
A WordPress.com site


